Bacterial strains and culture conditions
All clinical strains used in this study were obtained from the previously characterised R-GNOSIS collection of over 28,000 rectal samples collected from patients admitted to the Hospital Universitario Ramón y Cajal in Madrid between the years of 2014 and 2016 (R-GNOSIS-FP7-HEALTH-F3-2011-282512)).
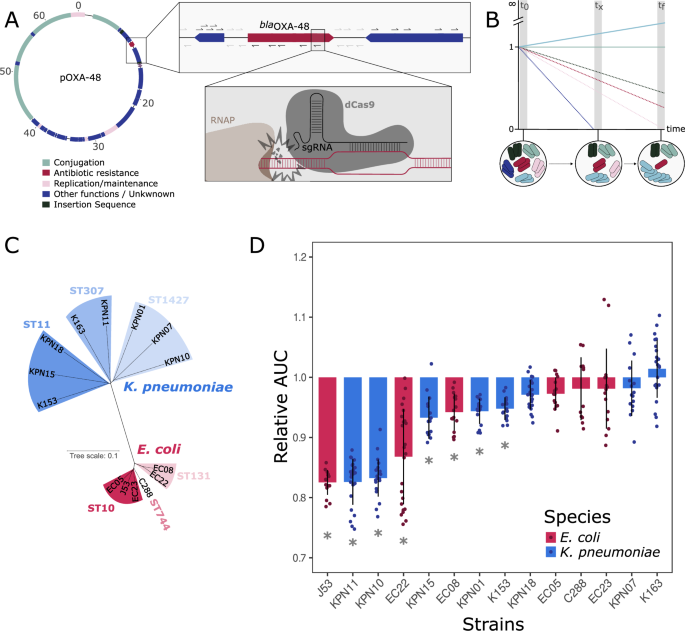
For the CRISPRi screens, a subset of these strains was selected based on previous works. First, a collection of strains that did not carry pOXA-48 naturally and where pOXA-48 transconjugants had been obtained37, and, secondly, a collection of strains that carried pOXA-48 naturally and where pOXA-48 had been successfully cured31. Out of these two collections, strains were selected based on sensibility to apramycin – antibiotic resistance gene used for selection of the OXAlib library -, conjugation rate efficiency, belonging to a sequence type of clinical relevance, their plasmid profile and, lastly, activity of dCas9 – tested by measuring cell death by silencing the essential gene rpsL, as in Cui et al38. and sfGFP (Supplementary Methods, SFig. 1, 2)). The resulting selection comprises 14 strains, 8 K. pneumoniae strains and 6 E. coli strains (including the lab strain J53) (Supplementary Data 2). The clinical strains belong to STs of clinical relevance, namely K. pneumoniae ST1427 (KPN01, KPN07 and KPN10), ST11 (KPN15, KPN18 and K153) and ST307 (KPN11 and K163), and E. coli ST10 (EC23), ST131 (EC08 and EC22), ST744 (C288) and ST4669 (EC05).
The variant of pOXA-48 used in this work is pOXA-48_K8 (accession number MT441554), and its sequence and complete genomes of the strains used in this study were obtained by the lab in previous works, available under the BioProjects PRJNA641166 and PRJNA803387 in the SRA repository of the National Centre for Biotechnology Information (NCBI).
All experiments were performed in Lennox lysogeny broth (LB), supplemented with 15 g/L of agar when indicated. The different antibiotics used throughout the study were amoxicillin / clavulanic acid at a proportion of 5 mg amoxicillin / 1 mg clavulanic acid, ertapenem, and apramycin. 50 µM of 2,4-diacetylphloroglucinol (DAPG) was used for induction of dCas9.
Calculation of pOXA-48 fitness effects in different clinical strains
The fitness effect associated with pOXA-48 in each strain was calculated by comparing differences in the Area Under the Curve (AUC) of isogenic plasmid-carrying and plasmid-free clones, as described in DelaFuente et al.58. Raw data can be found in the supplementary materials (Supplementary Data 4).
Designing the OXAlib library
dCas9 is reprogrammed to target a specific position using guide RNAs (sgRNAs), the design of which needs to follow a specific set of rules in bacteria59. (i) First, it has been shown that the specific sequence of the sgRNA has an effect on the activity of dCas9. (ii) Due to design limitations intrinsic to the CRISPRi technique, sgRNAs targeting coding regions have to be limited to the coding strand. The rules for efficiently targeting non-coding regions of DNA are not known, and therefore sgRNAs targeting those regions should be designed to target both strands of DNA. (iii) If dCas9 is directed to a gene within an operon, this binding will have a polar effect on the downstream genes. (iv) It is also important to avoid potential off-targets resulting from a permissiveness of dCas9 to bind to similar regions of DNA to bona fide targets.
The OXAlib library was designed using the CRISPR@Pasteur design tool (https://crispr-browser.pasteur.cloud/) which is trained on E. coli activity data, and which has been successfully used to design such libraries in different species of Enterobacterales59. The tool was adapted to our setup (looking for targets on a plasmid and for off-targets on a set of genomes – see Code Availability). Using this tool, the 5 best-performing sgRNAs were selected for each gene in pOXA-48_K8 (called pOXA-48 in this paper). sgRNAs targeting both DNA strands were also designed against intergenic regions of the plasmid (up to 5 targeting each strand, depending on the size of the intergenic region). The logic behind targeting both strands in intergenic regions is to attempt to capture effects of targeting any possible genetic elements present in the regions. 20 non-targeting control sgRNAs were also designed, amounting to a total of 568 sgRNAs (Supplementary Data 1).
Guides were selected with the previously described sorting strategy and ordered for synthesis as ssDNA oligos from Twist Bioscience. The sequence of each individual oligo is as follows: 5’-ACAGCTAGCTCAGTCCTAGGTATAATACTAGT-(20nt guide sequence)-GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGT-3’.
Cloning the OXAlib library
The ssDNA oligo pool was amplified using primers AC74 and AC75 (Supplementary Data 2) for subsequent cloning into pFR56apm26. pFR56apm is based on pFR5660, with an apramycin resistance gene as a selection marker. This antibiotic permits selection of the vector in all the different clinical strains (which due to their plasmidsome are naturally resistant to several antibiotics). Amplification was performed using New England Biolabs’ Phusion High-Fidelity DNA polymerase, with the HF buffer, DMSO and the recommended protocol. To minimise PCR amplification biases of specific oligos in the pool, the reaction was limited to 9 cycles (1 cycle for generating dsDNA oligos and 8 cycles of amplification), and performed with 1 ng of template and 50 pmol of each primer. The resulting product was gel purified. The pFR56apm vector was digested with BsaI and gel purified. The OXAlib plasmid library was assembled using the Gibson Assembly method61, using New England Biolabs’ NEBuilder HiFi DNA Assembly, 200 ng of total DNA and a ratio of 1:5 vector:insert. Cloning was performed in E. coli MG1655. Quality control of the final OXAlib library can be found in the Supplementary Methods.
Conjugating the OXAlib library into clinical strains
pFR56apm is a mobilisable plasmid – meaning it can be conjugated to compatible hosts, but requires expression of the conjugation machinery in trans. The OXAlib library was minipreped from E. coli MG1655 and transformed (with a coverage > 1000x of OXAlib) into E. coli ß3914ø + pTA-MOB62,63. This strain is auxotrophic for diaminopimelic acid (DAP), allowing for its counter-selection as a donor strain.
Conjugation of the OXAlib from ß3914ø to the different strains was performed by mixing 108 donor cells with 108 of recipient cells (ensuring a coverage > 1000x of OXAlib) for 3 h. After conjugation, the cells were selected in apramycin (50 µg/mL) and let grow overnight at room temperature. Absence of DAP counter-selects against ß3914ø. Conjugation efficiency was assessed using serial dilutions and a good coverage of the OXAlib library achieved for each strain (Supplementary Data 1).
CRISPRi screens
Experiment
First of all, it is crucial when working with CRISPRi screens to ensure that each step guarantees a good coverage of the library (i.e., that no individual guides are lost from the population due to over-dilution). To ensure this, we always work with an excess of cells of at least 1000x the size of OXAlib.
Strains carrying the OXAlib library were diluted 1:1000 in fresh LB / LB supplemented with ertapenem (subinhibitory concentrations of the antibiotics were used, Supplementary Methods, Supplementary Data 1). After 1 h, DAPG (50 µM) was added to the cultures for dCas9 induction. Every 8 h, a 1:1000 dilution was performed in fresh LB + DAPG / LB + DAPG + ertapenem. Prior to each dilution, OD was measured (to calculate number of generations). At 24, 48 and 72 h, apart from the dilution and OD measurement, a miniprep was performed using 10 mL of culture. The resulting pool of plasmids correspond to the timepoints at ~25, ~55 and ~85 generations. All screens were performed in triplicate (3 biological replicates), in aerobic conditions, at 37 °C, with shaking (250 r.p.m.).
Simultaneously, to assess whether the CRISPRi machinery encoded in pFR56apm is still active after 85 generations of growth, we grew the different strains harbouring pOXA-48::sfGFP and pFR56apm loaded with an sgRNA targeting sfGFP for 85 generations with induction of the CRISPRi machinery (DAPG 50 µm), then diluted the cultures in LB / LB supplemented with DAPG, grew the resulting sub-cultures overnight and measured sfGFP expression using cytometry. Our results suggest that the CRISPRi machinery is fully active until the end of the CRISPRi experiments performed in this manuscript (SFig. 16).
Illumina sample preparation and sequencing strategy
Library sequencing was performed as previously described38, with some small adaptations. The PCR primers used for the nested PCR can be found in the supplementary materials (Supplementary Data 2). To maintain good coverage of the OXAlib library, 100 ng of plasmid miniprep were used as template for each reaction. To minimise PCR biases, 19 cycles were used in the 1st PCR, and 13 cycles on the 2nd. Each replicate was given a different i5-i7 index combination, achieved by amplification with specific pairs of primers. Lastly, Illumina sequencing quality depends strongly on the variability of the regions sequenced. To bypass variability issues arising from amplicons sharing a large 5’ identical sequence, 3 different staggered primers were used in the 1st PCR.
Each sample was quantified using a bioanalyzer. 150 ng of each sample were pooled together and sequenced using the standard protocol of a NextSeq 500 benchtop sequencer (Illumina).
Data analysis
Quality control of Illumina sequencing
First, a QC of the raw Illumina data sequences (55 bp amplicon sequencing) was performed using FastQC v.0.11.964 and MultiQC v.1.1165. After confirming high quality per base score in the guide region (mean Phred score > 30), the sequences were used in downstream analyses.
Guide count per sample
For getting each individual guide count in each sample, each fastq file was parsed using Bash (see Code Availability). Briefly, knowing the sequence of the promoter preceding the guides in each read, 15 bp of it were used to match the reads position in which the guide began, and the following 20 bp (i.e., the sgRNA) were extracted. For each sample, the number of times each individual guide appeared was calculated using Bash commands. The list of guides was sorted, each unique occurrence was counted, and the output was reformatted to save each strain guides count in an individual csv file (see Source Data). Within the individual csv counts files, the guides with less counts than 5 for timepoints 0 and with less counts than 2 for the rest of the timepoints were filtered. The results of all the samples were merged in the same table, and only those guides present in the plasmid library were kept. The sequencing coverage for each guide was calculated as the median of the counts of guides (1 sgRNA per read; avoiding outliers from guides counted in excess) multiplied by 5 (as there are 5 guides per gene). The counts for the three replicates per time point were merged after checking the reproducibility of the replicates in each condition (LB + DAPG or LB + DAPG+ertapenem).
Gene score calculation
For calculating the gene scores (median log2 fold change by gene) a script which took the table with the replicates merged by sample, and calculated the log2FC for each gene, then centreing the values by removing the median of control guides, was written based on the code of Rousset et al.60 (https://gitlab.pasteur.fr/dbikard/ecocg/-/blob/master/Notebook.ipynb).
Phylogenetic analysis
The phylogenetic distances between the different strains were calculated using Mashtree v.1.2.066 (NJ method) and visualised using the iTOL web server (https://itol.embl.de/). Phylogenetic information was used to represent the strains clustered by relatedness in the heatmap (Fig. 2A).
Conservation of CRISPRi screening results in different strains
To analyse whether the effect of the OXAlib library on the strains tested was conserved, we studied the similarity between strains gene scores at the end of the screening. We calculated the correlation values (Pearson correlation coefficients) between the strains based on all the gene scores at the end of the screening for each condition by using the corrplot package v.0.95 in R. Additionally, we compared the genes whose silencing had the most notable effect in each strain. To do so, we listed the significant genes and intergenic regions for each strain and condition calculated from individual permutation tests for either LB + DAPG or LB + DAPG+ertapenem (see following Methods section). With these, we measured the Sørensen-Dice Similarity Coefficient (SDC) for all pairwise comparisons between strains of the same species screened in the same condition. The SDC was calculated using the following formula:
$$S{DC}=\frac{2{{{\rm{\cdot }}}}|{G}_{x}{\,\cap G}_{y}|}{|{G}_{x}|+|{G}_{y}|}$$
Where |Gx| and |Gy| are respectively the number of significant genes in strain X and Y (i.e. the number of elements compared from each strain), and |Gx ∩ Gy| is the number of common significant genes between X and Y strains in the given condition (i.e. full set of significant genes for both strains in either ertapenem or no antibiotics).
Permutation tests for gene importance
For checking statistically significant differences from the score of each gene from the distribution of gene scores in the CRISPRi screen, a permutation test for all the strains considered together per each condition (LB + DAPG or LB + DAPG + ERTA) was carried out. Briefly, the mean of each gene score was calculated considering all the strains from the screens (n = 14). The difference of this value and the mean of the scores from the rest of the group was assessed as the statistics of interest in the permutation test. The same statistic was calculated for 100000 iterations (i.e., minimum computable P value = 0.00001) taking random samples (n = 14) in each iteration. This number of permutations was selected to get more precise probabilities, as the P values of some genes resulted marginally significant when performing the permutation tests with fewer repeats (1000−10000). Lastly, the number of times that the permuted statistic was more extreme than the mean of each gene (i.e., the P value) was calculated. The same test was conducted for each species, as well as for each strain, for both experimental conditions. Finally, the p-values were adjusted by FDR. To report the statistical results from the permutation tests as well as the CRISPRi screening results, Volcano Plots were represented to simultaneously show each gene P value and its score using the R package EnhancedVolcano v.1.18.0. As in this type of screen, most genes showed scores close to 0. Thus, a log2FC low threshold (i.e., +−0.5) was set, close to the centre of the distribution. For the P value threshold, the default one (10e-06; i.e., P value < 0.05) was selected.
Ridge regression model for plasmid cost
To simultaneously consider the contribution of multiple genes to pOXA-48 cost, a multiple regression model with Ridge regularisation was developed using the glmnet v.4.1-7 R package separately for both E. coli and K. pneumoniae. First, feature selection of all the genes and intergenic regions tested was performed, as there were 8 observations of plasmid cost for K. pneumoniae and 6 observations of plasmid cost for E. coli (n=strains), and 106 explanatory variables (n=genes & intergenic regions included in CRISPRi screen). Then, only those genes with significant P values in the permutation test performed for each of the species (i.e., expected to be more informative) in absence of antibiotics during the CRISPRi screen were included in the model. From these, intergenic regions were kept out, as their effect introduced multicollinearity issues by showing similar behaviour as their surrounding genes in the CRISPRi screening (e.g., blaOXA-48 intergenic surrounding regions). Multicollinearity between explanatory variables was assessed after feature selection using the R package corrplot v.0.95. Due to the limited number of observations for each species, the models were trained using Leave-One-Out Cross Validation. Moreover, to include all the genes resulting from the feature selection process, Ridge regularisation was selected, as the regression coefficients are not shrunk to 0. Optimal parameters from the trained models were selected to then build the final models. The ridge coefficients for each gene, and the R-squared for each species were calculated from the final models tuned with the optimal parameters (lambda (λ); SFig. 8).
Plasmid cost based on guide distribution
To infer the plasmid cost in each strain, a proxy based on the general effect of blocking the plasmid genes was employed. The effect of each plasmid gene on bacterial fitness is expected to be reflected on the CRISPRi screen in absence of antibiotics, as guides which block costly genes that cause deleterious effects are expected to show higher scores, and vice versa. Then, if the plasmid is highly costly in a strain, the general effect of blocking its genes should reflect this. Thus, the general effects of blocking the plasmid genes were correlated with the relative fitness of plasmid-carrying strains. The difference between the mean effect of blocking pOXA-48 genes (mean CRISPRi gene scores for each strain) and the mean effect of the control guides, which are supposed to have no effect, was calculated. Then, these values were correlated (Spearman method) with the fitness effect of pOXA-48 in each strain.
Validation of CRISPRi screen’s results
Strain selection
From the initial set of strains used in this study, two E. coli (C288, EC22) and three K. pneumoniae strains (KPN10, KPN11 and KPN18) were chosen for validation of results based on the overall strongest fold-changes observed.
Cloning of individual sgRNAs in pFR56apm and conjugation into clinical strains
Cloning of individual sgRNAs (Supplementary Data 1) in pFR56apm was performed using Golden Gate assembly as in Rousset et al.60. All cloning was performed in E. coli MG1655 and followed by conjugation into the clinical strains as described previously.
blaOXA-48 associated fitness effects
To validate the results associated with silencing the gene blaOXA-48, growth curves were performed to study plasmid fitness effects as explained before in this method section. This time, a second set of strains was selected from the RGNOSIS collection, to avoid overfitting to the original strains used in the screen. These strains were once again selected based on the fitness effects associated with the plasmid, as described in Fernández-Calvet et al.16. A variant of pOXA-48 lacking a complete blaOXA-48 was identified previously in the lab31. This variant was conjugated on the cured version of the strain, obtaining three isogenic set-ups for each strain: the wild-type carrying pOXA-48, a pOXA-48-free cured version, and a pOXA-48∆blaOXA-48 transconjugant.
Plasmid stability
Plasmid stability of pOXA-48 and pOXA-48 versions was measured by growing cells carrying the plasmid in LB over subsequent periods of 24 h. After each cycle, cells were plated in LB, guaranteeing the isolation of individual colonies. Cells were then streaked in presence and absence of ertapenem using replica plating. Percentages of plasmid loss / plasmid presence were calculated by comparing growth in presence and absence of selection. Each experiment was performed in triplicate. Number of 24 h cycles used in each experiment are indicated throughout the text. To statistically analyse whether the stability of pOXA-48 was significantly affected in at least one strain when silencing individual genes (Fig. 3B), we performed a one-tailed z-test for each guide. Briefly, we calculated the maximum deviation from the control group (guide “control”) per individual guide, from which we inferred their Z-score. Finally, we corrected the P values by Bonferroni adjustment (n = 8).
qPCR for PCN calculation
Changes in pOXA-48’s PCN were calculated for the different clinical strains carrying pFR56apm programmed to silence different plasmid genes. In order to do so, the different strains were grown in triplicates in LB supplemented with apramycin 50 µg/mL and DAPG 50 µM for 24 h. 50 µL samples were taken of each replicate, centrifuged to collect the pellet and boiled for 10 min. 2 µL of each boiling reaction were used per qPCR reaction, using the NZYSupreme qPCR Green Master Mix (2X), ROX plus kit from NZYTech. Targeted plasmid and chromosome genes were blaOXA-48 (amplicon size 100 bp; efficiency 107,47%) and dnaE (chromosomal gene with 1 copy, amplicon size 200 bp, efficiency 105.79%), respectively. Efficiency was calculated (taking into account amplicon size difference) as in San Millán et al.67. The amplification conditions were: 5 min of initial denaturation (95 °C), followed by 30 cycles of 15 s denaturation, 30 s annealing (55 °C) and 30 s extension (60 °C). The relative PCN was calculated as in San Millán et al.67. Raw data for qPCR can be found in the supplementary data (Supplementary Data 4).
Lambda-Red recombination for construction of deletion mutants
A version of pOXA-48 lacking pemK was built using lambda-red recombination68. The recombination cassette with extremes matching the beginning and end of pemK, and a kanamycin gene flanked by FRT sides was amplified from pKD4, as described in Datsenko et al.68. Primers used for construction of the cassette and validation of the construct can be found in the supplementary data (Supplementary Data 2).
Statistics & reproducibility
All statistical analyses were performed using RStudio v.2024.12.1 + 563 and R v.4.3. Parametric or non-parametric tests were used depending on the structure of the data (see each specific subsection analysis). Tests were controlled for multiple comparison testing by applying FDR or Bonferroni adjustment (see each specific subsection analysis). To analyze correlation data, we used Pearson correlation for normally distributed data. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomised. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.