Preparation of genotype–fitness dataset
We retrieved all SARS-CoV-2 genome sequences and their associated metadata available as of November 2, 2023, from GISAID. We then assigned the most recent PANGO lineage classification available at that time to each sequence in our dataset using Nextclade v.2.14.046 with Nextclade dataset version “2023-10-26T12:00:00Z.” Then we excluded low-quality sequences based on the following criteria: (i) absence of collection date information; (ii) samples derived from animals other than humans; or (iii) more than 1% undetermined nucleotide characters.
To develop a classification system for SARS-CoV-2 with a higher resolution than the PANGO lineage and based solely on the sequence of the S protein, we defined the genotype of the S protein and utilized it as the virus classification system in this study. The S protein genotypes refer to groups of viral sequences that share a unique set of mutations in the S protein. To achieve this, we first identified mutations in the S protein observed in more than 100 sequences. We then analyzed the mutation patterns across each sequence, enabling us to categorize sequences into genotypes based on these patterns. Only genotypes represented by 20 or more sequences in any country were considered for our analysis. As a result, a total of 13,643 S protein genotypes were included in our dataset. Each genotype was linked to the Nextclade PANGO lineage, Nextclade clade, and a representative genome sequence. This representative sequence was randomly chosen from the collection of sequences of the genotype. The emergence date of a genotype was determined as the 1st percentile date of collection for the viral isolates within the genotype. The viral genome sequences contained within this dataset are summarized under the EPI_SET_ID: EPI_SET_240311ma, which can be accessed through the GISAID website (https://gisaid.org/).
To estimate the relative Re value of each genotype in each country, we began by tallying the daily count of each genotype within each country’s dataset. We applied a multinomial logistic model to the count data of each country as previously described8. In this model, time (date) was used as the explanatory variable and the detection count for each variant as the dependent variable. Parameters in the model were estimated using the maximum likelihood method with the “multinom” function in the “nnet” package v.7.3.18 in R v.4.2.1.
In estimating Re within the multinomial logistic model, it is necessary to select one variant as the reference variant with Re = 1. Ideally, the reference variant should have the largest possible number of sequences. Additionally, a common reference variant must be chosen for all countries included in the analysis. To do this, we first counted how many sequences of each variant were detected in each country. Then, we compared the detection counts across countries for each variant and calculated the minimum count for each variant across countries. Finally, the variant with the highest minimum count was selected as the common reference variant for all countries. This approach allows us to choose a reference variant with the largest number of sequences registered across all countries, or at least in the country with the fewest sequences. Accordingly, the major genotype of the BA.5 lineage (equivalent to BA.5.2.1 in the PANGO lineage) was selected as the reference.
We then extracted the estimated growth rate (slope parameter) of each genotype relative to a reference genotype. The relative Re of the viral lineage l, \({r}_{l}\), was calculated according to the slope parameter \({{{\rm{\beta }}}}_{l}\) of the lineage as \({r}_{l}=\exp \left({{\rm{\gamma }}}{{{\rm{\beta }}}}_{l}\right)\), where \({{\rm{\gamma }}}\) is the average viral generation time (2.1 days) (http://sonorouschocolate.com/covid19/index.php?title=Estimating_Generation_Time_Of_Omicron)7. The estimated Re value for each genotype is summarized in Supplemental Data 1.
Estimating relative Re with the multinomial logistic regression model has certain limitations. The first is that it can only estimate relative Re values between variants; in other words, it cannot provide absolute (non-relative) values of Re. As a result, we cannot determine solely based on these values whether an epidemic caused by a particular variant will expand or subside. In this model, a variant’s disappearance from the viral population (its relative frequency becoming negligible) is driven only by the expansion of other variants with a higher Re. The second limitation of this model is the assumption that the relative Re between variants remains constant over time. In reality, the relative fitness of variants can fluctuate in response to changes in the host population’s immune status, influenced by factors such as natural infections and vaccinations. In 2024, when most of the population has some immunity to SARS-CoV-2 due to vaccination or prior infection, the impact of immune escape on fitness is likely more pronounced compared to the early stages of the COVID-19 pandemic in 2020. The third limitation of this model is the assumption that the average viral generation time remains constant across variants. This assumption does not always hold, as previous studies have estimated differences in generation time between the Delta and Omicron variants47. Therefore, it is essential to interpret the estimated relative Re values with these limitations in mind.
In each viral clade, a small number of viral genotypes had emergence dates that were exceptionally earlier than the majority of other genotypes. For instance, the earliest emergence date in clade 21 K (BA.1) is associated with hap_89965, which is recorded as January 1, 2021. However, considering that BA.1 was first identified in November 2021, this date is likely erroneous. Additionally, in the analysis of prediction performance for future variants described later, it is preferable for the past–future cutoff dates (set at 1-month intervals) to align as closely as possible with the actual emergence dates of the viral clades. Therefore, we excluded a small number of these earliest genotypes with anomalously early emergence dates from the genotype–fitness dataset. Specifically, genotypes with emergence dates earlier than November 30, 2021 for clade 21K (BA.1); December 31, 2021 for clade 21L (BA.2); March 31, 2022 for clades 22A (BA.4) and 22B (BA.5); January 31, 2022 for clade 22C (BA.12.1); May 31, 2022 for clade 22D (BA.2.75); August 31, 2022 for clades 22E (BQ.1) and 22F (XBB); November 30, 2022 for clade 23A (XBB.1.5); February 28, 2023 for clade 23B (XBB.1.16); September 30, 2022 for clade 23C (CH.1.1); and May 31, 2023 for clade 23F (EG.5.1) were removed from the dataset.
Prior to the training step, we excluded S protein sequences containing more than 5 ambiguous characters and more than 30 amino acid deletions from the dataset. Furthermore, we excluded viral genotypes classified as Recombination clades (i.e., recombinant lineages, excluding major recombinant lineages such as XBB) in the Nextclade classification. Also, we focused on countries where more than 300 genotypes were detected, which led to collecting data for 17 countries: Australia, Belgium, Brazil, Canada, Denmark, France, Germany, India, Italy, Japan, Netherlands, South Korea, Spain, Sweden, Switzerland, the UK, and the USA. As a result of this additional filtering, a total of 21,281 genotype–fitness (relative Re) data points, encompassing 12,817 genotypes across 17 countries, were included in our genotype–fitness dataset.
The estimated fitness value was transformed using the natural logarithm function, and then the data was scaled so that the 0.1 percentile and 99.9 percentile points fall between 0 and 1 before model training.
Preparation of DMS data for mAbs evasion
In this study, we utilized DMS data on evasion from mAbs provided by Cao et al.24. The processed DMS data, specifically the mutation-wise immune escape score prepared for the antibody-escape estimator developed by Greaney et al.37, was downloaded from the Bloom lab GitHub repository on April 11, 2023 (https://github.com/jbloomlab/SARS2_RBD_Ab_escape_maps/blob/main/processed_data/escape_data_mutation.csv). We applied specific exclusion criteria to the DMS data: (i) mAbs categorized as “SARS convalescents” and “WT-engineered”; and (ii) mAbs with an IC50 value ≥ 10, indicative of very weak binding affinity, for the target virus. The escape score in this repository was calculated using a DMS experiment using the ancestral D614G strain’s RBD. Following the methods of Greaney et al., we defined a weighted escape score for each target virus (e.g., D614G and BA.2) from this escape score, following the method of Greaney et al. Specifically, the escape score was multiplied by the IC50 value for the S protein of the target virus, followed by negative log transformation with a pseudo count of 1. The weighted escape score was scaled so that the 0 and 95 percentiles fell within the range 0–1, and values above 95 percentile were clipped to 1. For the comprehensive training of CoVFit, the weighted escape values for D614G were employed. On the other hand, considering that variants predominantly circulating after early 2022 are related to the BA.2 lineages, the weighted escape values for BA.2 were used in the training for CoVFitPast.
Dataset preparation for domain adaptation
The S protein sequences for Coronaviridae, except for SARS-CoV-2, were downloaded from the NCBI Identical Protein Groups database (https://www.ncbi.nlm.nih.gov/ipg) on July 3, 2023, using the following search query: query:(“Alphacoronavirus”(Organism) OR “Betacoronavirus”(Organism) OR “Gammacoronavirus”(Organism) OR coronavirus(All Fields)) AND (spike(All Fields) OR S(All Fields) OR surface(All Fields)) NOT (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR (“Severe acute respiratory syndrome coronavirus 2”(Organism) OR SARS-CoV-2(All Fields))))))))))) NOT (“unidentified”(Organism) OR (“unidentified”(Organism) OR (“unidentified”(Organism) OR (“unidentified”(Organism) OR (“unidentified”(Organism) OR “Unknown”(All Fields)))))) NOT “unidentified human coronavirus”(Organism) NOT (“synthetic construct”(Organism)) AND (“1000”(SLEN): “1500”(SLEN)). The metadata associated with these sequences were also downloaded. The S protein for the SARS-CoV-2 Wuhan-Hu-1 strain was downloaded using NCBI Datasets Command-line tools v.15.6.1 (https://www.ncbi.nlm.nih.gov/datasets/docs/v2/download-and-install/) and subsequently incorporated into our dataset. Sequences with more than 5% unidentified amino acids were filtered out. Next, we removed redundant sequences using CD-HIT v.4.8.148 with a clustering threshold of 99% sequence identity. However, even after the CD-HIT filtering, a large number of sequences (i.e., 796 sequences) corresponding to the porcine epidemic diarrhea virus (PEDV) remained in the dataset. To reduce the redundancy of this virus group, we randomly selected 10 representative PEDV sequences to retain in our dataset. Consequently, 1392 Coronaviridae sequences were included in our dataset.
In addition to the Coronaviridae S protein dataset, we prepared a dataset for the SARS-CoV-2 S protein specifically for domain adaptation. Of the S protein genotypes we defined in this study (see the section “Preparation of genotype–fitness dataset”), we eliminated genotypes with an emergence date later than August 31, 2022, in order to prevent the model from accessing data beyond this cutoff date during the domain adaptation process. Subsequently, we removed redundant sequences using CD-HIT with a clustering threshold of 99%, in accordance with the method described above. Consequently, 114 S protein sequences of SARS-CoV-2 were retained in the dataset. Finally, we combined the Coronaviridae and SARS-CoV-2 S protein datasets, resulting in 1506 sequences, for use in domain adaptation. The S protein sequence information used in the domain adaptation step is summarized in Supplemental Data 2.
Introduction of CoVFit
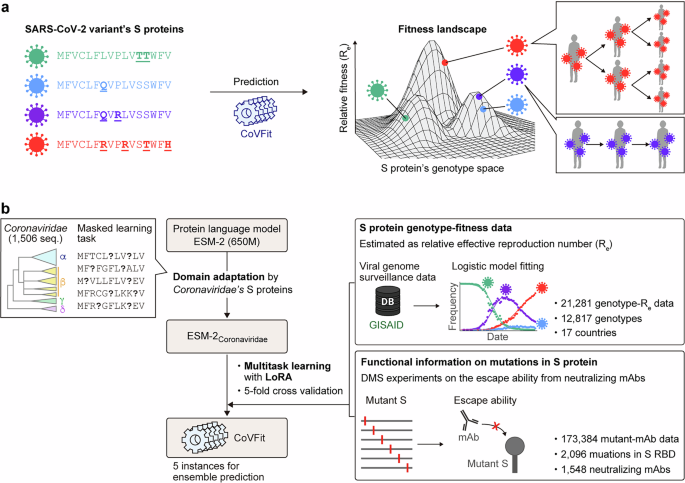
We developed CoVFit, a fitness prediction model based on S protein sequences, by finetuning the ESM-2 protein language model (Fig. 1a). To enhance the model’s performance, we employed three key techniques: domain adaptation, multitask learning, and Low-Rank Adaptation (LoRA)49. Domain adaptation is an additional pretraining phase using a custom data collection. In this study, we performed domain adaptation using S protein sequences from various human and animal coronaviruses (see the “Dataset preparation for domain adaptation” section). This technique enabled the model to better learn the general properties of these proteins (see the “Domain adaptation” section). Multitask learning, a framework that trains a model on different types of data simultaneously, was utilized to allow the model to capture critical information shared across tasks, thereby enhancing its generalization capabilities. In CoVFit, the model was finetuned using a total of 1,565 regression tasks, including to predict fitness values for 17 countries and to predict relative binding affinity for 1548 mAbs (see the “Model architecture of CoVFit” section). Finally, LoRA is a technique to fine-tune large models efficiently that reduces the requirements for GPU memory resources without compromising the model’s prediction performance. LoRA additionally contributes to mitigating the model’s tendency to overfit (see the “Model finetuning and performance evaluation” section for further information).
The input for CoVFit consists of amino acid sequences of SARS-CoV-2 S proteins, aligned with the S protein of the Wuhan-Hu-1 strain. These aligned sequences can be generated by Nextclade. CoVFit can predict the fitness (relative Re) value across 17 countries and the ability to evade 1548 types of mAbs for a given S protein sequence (Fig. S1a).
Training of the CoVFit model completes within 24 h on a computational node with a single Nvidia A100 GPU (40GB) for each instance. Consequently, the model can be updated routinely using the latest genome surveillance data without intensive computational resource requirements.
Utilizing a five-fold cross-validation scheme, we generated five instances of the CoVFit model, which enabled us to estimate both the average prediction value and its uncertainty across these models (Fig. 1b). This approach was chosen because the predicted values, especially regarding the fitness of future variants, can vary among different instances of the trained models (Fig. 3).
CoVFit implementation
ESM-2 models with various parameter sizes are available32 (https://github.com/facebookresearch/esm). Of these models, we used the version with 650M parameters, prioritizing a balance between prediction performance and computational cost. According to the official benchmark using the unsupervised contact prediction task, this 650M parameter model achieves a performance 1.7 times superior compared to the 35M parameter model, which possesses 20 times fewer parameters. However, the performance gain when comparing the 650M model to the 15 billion (B) parameter model, which has 20 times more parameters, is relatively modest at only 1.08 times. Furthermore, a systematic analysis presented in a recent preprint indicates that enlarging the parameter size of a protein language model does not necessarily enhance prediction performance for tasks outside of protein structure prediction50. Given this insight, we opted not to employ models larger than the 650M model, such as the 3B or 15B models, in our study.
The ESM-2 model has a maximum input sequence length (1024 amino acids) due to the computational demands of self-attention, which requires memory in proportion to the square of the sequence length (O(L2)). Unfortunately, the S protein of the Wuhan-Hu-1 strain is composed of 1273 amino acids, exceeding the model’s limit. Consequently, amino acid sequences beyond the 1024th position (amino acids 1025–1273; the C-terminus of the S2 subunit) are truncated and not utilized in the ESM-2 model. This constitutes a technical limitation of CoVFit. Nonetheless, this limitation is anticipated to minimally impact performance, considering that while mutations predominantly occur within the S1 subunit (amino acids 1–681), the S2 subunit (amino acids 682–1273) remains highly conserved and with fewer mutations.
CoVFit was implemented using Python v.3.11.4, NVIDIA CUDA v.12.1.0, PyTorch v.2.1.0, Transformers v.4.31.0, and PEFT v.0.5.0. Further information about the system requirements for CoVFit can be found in the GitHub repository (https://github.com/TheSatoLab/CoVFit). The computational codes were executed on a supercomputer node equipped with a single NVIDIA A100 GPU with 40 GB RAM unless otherwise noted.
Domain adaptation
To establish the ESM-2Coronaviridae model, domain adaptation was carried out using the masked language learning scheme as described in Delvin et al.51. For domain adaptation, the S protein dataset prepared in the “Dataset preparation for domain adaptation” section was used. For our model’s domain adaptation training, each input sequence had 15% of its positions masked randomly, with each instance of a position’s masking having an 80% chance to be a token, a 10% chance to be incorrect, and a 10% chance to be the original. Subsequently, amino acid or token types for these 15% of positions were predicted in batched training steps, and model weights were updated using a cross-entropy loss function.
Using the scheme above, we trained the 650M parameter ESM-2 model with the provided MaskedLM layer, downloaded via functions implemented in the Hugging Face Transformers library. The model was trained for 30 epochs. The batch size was set at 5. A base learning rate of 2e\(-\)5 was used with one epoch of warmup, and a cosine-based learning rate scheduler was implemented to successively lower the learning rate during training.
To compare the inference ability of the ESM-2Coronaviridae model to the original ESM-2 model, we performed inference with both models on masked SARS-CoV-2 S protein sequences. Since our dataset for domain adaptation training includes the S proteins of genotypes that emerged up to August 31, 2022, we used genotypes with emergence dates later than September 1, 2022, for inference. The same masking parameters as in the training were used. The results on the test dataset were converted to perplexity scores as the exponential of the cross-entropy loss value calculated during inference. Given as \({perplexity}=\,{{{e}}}^{-{\sum }_{x}P\left(x\right)\log Q(x)}\) where \(P(x)\) is the true probability distribution and \(Q(x)\) is the probability distribution from the model’s predictions, the perplexity score represents how certain the model is in making its predictions, with lower values demonstrating higher certainty. For our inference results, the original ESM-2 model produced a perplexity score of 11.38, whereas the ESM-2Coronaviridae model achieved a low perplexity score of 1.17, demonstrating higher prediction certainty after domain adaptation training (Fig. S1c).
To assess the possibility of the domain adaptation negatively impacting the model’s original ability to provide inference on a wide variety of proteins, we again compared the original EMS-2 and ESM-2Coronaviridae models, this time with protein sequences sampled from the UniRef50 released in March 2018. A subset consisting of 29,950 sequences was randomly sampled from the full 3,016,211 sequences and used for the evaluation. The perplexity values of the models were checked as above for the two models, with the original ESM-2 model’s perplexity score at 6.76 and the ESM-2Coronaviridae model’s perplexity score at 6.86, demonstrating that the model retains its certainty on general proteins after domain adaptation (Fig. S1c).
We conducted the domain adaptation training in Python v3.10.9 with CUDA v12.1.1 and torch v2.1.0.dev20230601 using the Hugging Face Transformers v4.34.1 library.
The computation was executed on a single NVIDIA RTX 6000 Ada GPU with 48 GB RAM. More detailed information on implementation is available in the GitHub repository (https://github.com/TheSatoLab/CoVFit).
Model architecture of CoVFit
For the multitask learning component, we engineered custom task-specific regression heads for the ESM-2 model (Fig. S1a). On the embedding layer of ESM-2, a linear layer with dimensions equal to the number of tasks was set as task-specific heads. Additionally, an intermediate linear layer with 252 dimensions connecting the embedding layer and the task-head layer was set.
In CoVFit, a single input sequence is linked to multiple target variables due to the multitask learning framework. For example, regarding DMS data, a typical S protein mutant (input sequence) is linked to relative binding affinity values for >1000 mAbs (target variables). To boost computational efficiency, CoVFit utilizes an architecture that processes a single input sequence alongside its multiple corresponding target variables in parallel, rather than processing pairs of the same input sequence and one target variable sequentially (Fig. S1b). As a result, the loss values for multiple target variables associated with a single input sequence are calculated simultaneously.
However, the number of tasks linked to each input sequence can differ greatly, especially when comparing the variant S protein sequences used for fitness prediction (up to 17 tasks) against the mutant sequences used for DMS predictions (up to 1548 tasks). Consequently, the magnitude of the loss value for each dataset can vary significantly based on the number of associated tasks, which can lead to training instability. To stabilize the training process, CoVFit utilized non-overlapping random sampling to create data chunks where a single input sequence is associated with target variables for a maximum of 10 tasks. These generated sequence-variable chunks were then used as the training inputs.
For the loss function, CoVFit utilizes a custom least squares approach weighted according to individual tasks. In principle, the weights were determined to be proportional to the reciprocals of the task frequencies. One exception was implemented where, for fitness prediction tasks for genotypes that emerged after January 1, 2022, we adjusted the weights by doubling them.
Model finetuning and performance evaluation
In CoVFit, we finetuned the custom model based on ESM-2 using the LoRA technique implemented in the Hugging Face PEFT v.0.5.0. Low-rank adapters were injected into the weight matrices of the key, query, and value components, as well as those for the dense layers. Full finetuning was applied to these adapters and the custom regression heads added onto ESM-2, while the other, original layers were kept frozen in their pretrained state. A rank parameter of r = 8 and a scale parameter of alpha = 16 were used. Consequently, out of the total 659,741,475 parameters, the model has 7,768,974 trainable parameters, which constitutes ~1.18% of the total. The LoRA dropout rate was set at 0.05.
For finetuning, the AdamW optimizer was used with a weight decay parameter of 0.02. The maximum learning rate was set at 2.0E-4 with a linear learning rate scheduler, and the training was conducted over 20 epochs with a warmup ratio of 0.05. The batch size was set at 4 with gradient accumulation steps of 2.
The genotype–fitness and DMS datasets were randomly divided into training, evaluation, and test datasets in a 6:2:2 ratio. For the genotype–fitness dataset, we considered the combinations of country and Nextclade clade, ensuring that data representing each combination were evenly distributed across the training-evaluation and test datasets. Similarly, for the DMS dataset, the types of mAbs were considered during the data splitting process. We conducted the data splitting with a five-fold cross-validation approach.
In our experiments aimed at assessing the model’s ability to predict the performance of future variants, we began by dividing the genotype–fitness dataset into two: one for past variants and another for future variants, based on their emergence dates. We generated eight datasets using eight cutoff dates, spaced 1 month apart, from January 31, 2022, to August 31, 2022 (Fig. 3b). Training was done using these datasets according to the scheme described above.
In our experiments designed to assess the importance of including DMS data for immune evasion, we trained alternative instances without incorporating the DMS dataset for comparison. Likewise, in our experiments evaluating the significance of the domain adaptation step, we employed the original ESM-2 model rather than the version adapted to the coronaviral S protein dataset.
CoVFit-CLI
The CoVFit-CLI tool packages CoVFitNov23 via pyinstaller 6.4.0 using Python v.3.10.9, torch v.2.1.2, transformers 4.37.1, and bio v.1.5.9 with CUDA v.12.3 on x86_64 Linux, kernel 5.15.0.
Development of fitness prediction models based on non-deep learning models
We constructed fitness prediction models based on LASSO, Random Forest, and LightGBM to compare the prediction performance of CoVFit with those of these models. LASSO employs a linear regression framework enhanced with L1 regularization, offering a method to include penalty terms that reduce overfitting by shrinking some coefficients to zero. In contrast, Random Forest and LightGBM are advanced, decision tree-based models known for their greater expressive capability. These models aim to predict a variant’s fitness value based on its amino acid mutation profile in the S protein and the country of origin. Both the mutation profile and the country data were one-hot encoded to serve as input features for the models.
We trained these models and evaluated their performance using the past–future variant datasets with eight cutoff dates, spaced 1 month apart, from January 31, 2022, to August 31, 2022, and the five-fold cross-validation scheme as described in the “Model finetuning and performance evaluation” section. In the training dataset, we selected 200 features to be used as inputs of the models according to the feature importance estimated by Random Forest. We trained the models with hyperparameter-tuning using a Bayesian optimization method. In this process, R2 was used as the optimization metric, and the number of iterations was set at 20. The parameter spaces searched in this step are described in detail in the GitHub repository (https://github.com/TheSatoLab/CoVFit).
The machine learning models above were reconstructed using Python v.3.9.13, pandas v.1.4.4, numpy v.1.21.5, lightgbm v.3.3.5, scikit-learn v.1.0.2, and scikit-optimize v.0.9.0.
Phylogenetic analysis
We created the dataset for phylogenetic analysis as a subset of the dataset of the representative viral genome sequences encoding respective S protein genotypes (EPI_SET_ID: EPI_SET_240311ma; https://www.gisaid.org; see “Preparation of genotype–fitness dataset” section). We removed sequences matching the following criteria: (i) sequences with >3% ambiguous characters across positions 265 to 29,673 (in alignment with the Wuhan-Hu-1 reference (GenBank accession number: NC_045512.2 (https://www.ncbi.nlm.nih.gov/nuccore/1798174254))) and (ii) sequences classified as “recombinant” according to Nextclade clade assignments. Additionally, we included the Wuhan-Hu-1 reference genome sequence to our dataset. The dataset for viral genome sequences used in the phylogenetic analysis, except for the Wuhan-Hu-1 reference genome, is summarized under the EPI_SET_ID: EPI_SET_240311rk, which can be accessed through the GISAID website (https://gisaid.org/).
The nucleotide viral genome sequences were aligned to the reference sequence of Wuhan-Hu-1 using Minimap2 v.2.1752. This alignment was then converted into a multiple sequence alignment following the GISAID phylogenetic analysis pipeline (https://github.com/roblanf/sarscov2phylo). Sites corresponding to positions 1–265 and 29,674–29,903 in the reference genome were masked, that is, converted to “NNN,” to exclude them from subsequent analyses. The maximum likelihood phylogenetic tree was constructed using IQ-TREE v.2.1.4_beta, adopting the GTR+I+G nucleotide substitution model53. To assess the reliability of the phylogenetic tree nodes, an ultrafast bootstrap analysis was performed with 1000 replicates. A time-resolved phylogenetic tree was inferred from the constructed tree using TreeTime v.0.11.1, with the rerooting strategy set to “oldest”54, resulting in rerooting by the Wuhan-Hu-1 strain. The S protein sequences for ancestral nodes were also reconstructed using TimeTree with the default options.
Detection of phylogenetic branches with fitness elevation using CoVFit
To infer the impact of mutations on fitness through the observed evolution of SARS-CoV-2, we analyzed the increase in predicted fitness across all branches of the SARS-CoV-2 phylogenetic tree, as outlined in the “Phylogenetic analysis” section. We employed five CoVFitNov23 models, developed via a five-fold cross-validation, to predict the fitness values for both existing and reconstructed ancestral S protein sequences within the tree. Since CoVFit predicts fitness across multiple countries, we averaged these predictions to obtain a single representative fitness value for each sequence, resulting in five representative fitness values per sequence. We compared these values between each node and its ancestral node, calculating the mean fitness gain for the branches connecting them. Statistical significance of fitness changes was determined using a paired Welch’s t-test, with multiple testing correction applied via the Benjamini–Hochberg method. Branches with an FDR less than 0.1 were considered statistically significant. We also identified mutations in the S protein acquired along each branch by comparing the S protein sequences at both ends of the branch. The detected fitness elevation events are summarized in Supplemental Data 3.
Characterization of the F456L substitution using publicly available DMS data
Position-wise scores for escape from humoral immunity were calculated using escape estimator37 based on DMS data for the ability to evade mAbs presented in Cao et al.24 (https://github.com/jbloomlab/SARS2_RBD_Ab_escape_maps) (shown in Fig. 5c). For the ACE2 binding and protein expression DMS data, we retrieved the per-site variant score results presented by Taylor and Starr36 (https://github.com/tstarrlab/SARS-CoV-2-RBD_DMS_Omicron-XBB-BQ/blob/main/results/final_variant_scores/final_variant_scores.csv). We filtered for mutant L on position 456 and retrieved the “delta_bind” and “delta_expr” values presenting the mean of values across replicates minus the mean for the reference residue for each variant target (shown in Fig. 5d).
CoVFit-based in silico (deep) mutational scanning analysis
Instances of the CoVFitnonJN1 model, models trained on the dataset without the sequences of JN.1, were utilized to infer the fitness of S protein mutants. First, the mean fitness value for each mutant was calculated across different countries. Subsequently, these mean values were averaged across all five CoVFitnonJN1 model instances, yielding a singular average fitness value for each S protein mutant. This streamlined fitness value was then compared to the fitness of the original backbone S protein sequence.
Epidemic analysis on JN.1 subvariants
We retrieved all SARS-CoV-2 genome sequences and their associated metadata available up to October 21, 2024, from GISAID. To ensure data quality, sequences were excluded from analysis based on the following criteria: (i) absence of collection date; (ii) samples taken from animals other than humans; (iii) more than 2% undetermined nucleotides; or (iv) samples collected during quarantine. We analyzed the BA.2.86 lineage collected between October 1, 2023, and July 31, 2024 (EPI SET ID: EPI_SET_241126wq).
We calculated the proportion of viral sequences harboring mutations at specific sites within the BA.2.86 lineage. Each viral lineage category includes its descendant lineages unless those descendant lineages are explicitly defined as separate categories. For the temporal trends in mutation and variant detection frequencies, calculations were performed at 7-day intervals for each geographic region within the BA.2.86 population. Results from Africa and South America were excluded due to the low total sequence count.
Methodological discussion: room for improvement of CoVFit
We recognize the presence of multiple areas where CoVFit could potentially be improved with future development. First, since our current model is solely trained on S protein sequences, it may be possible to improve its performance by including information on additional viral proteins. Previous studies have identified mutations associated with increased fitness also in non-S proteins, particularly in the nucleocapsid (N) protein, supporting the possible effectiveness of this approach4. In the current setting, the effects of mutations in non-S proteins are absorbed into the effects of mutations in the S protein that have linkage disequilibrium relationships with these mutations. However, it is certain that the S protein has a particularly strong impact on fitness compared to other viral proteins4. Therefore, it is unclear to what extent prediction accuracy would be improved by adding information from other viral proteins. There is even a possibility that generalizability could be decreased by including other viral proteins due to the decrease in signal-to-noise ratio. Similarly, although the amino acid sequences of the S protein beyond the 1024th position (amino acids 1025–1273; the C-terminus of the S2 subunit) are truncated and not utilized in the ESM-2 model, it is unclear whether this limitation has a negative effect on prediction performance, as the S2 subunit (amino acids 682–1273) remains highly conserved and has fewer mutations.
Second, it may also be possible to improve the performance by including various DMS data, such as those on other viral phenotypes. In this study, we only used DMS data on the immune evasion ability against mAbs. However, given the significant impact of the ACE2-binding affinity of the S protein on fitness, employing DMS data for this trait could improve predictive performance. In our preliminary experiments, however, we found that the convergence speed for DMS data on binding affinity to ACE2 was much slower compared to other tasks. Considering the difficulty of simultaneous learning, we decided not to use this DMS data in this study. Similarly, while we used DMS data based on the ancestral strain (D614G), incorporating DMS data obtained by experiments using S proteins of other variants (particularly variants emerged recently) could potentially further improve predictive performance.
The third consideration is the scaling up of the model. It is generally known that language models improve in performance as they increase in size. Indeed, benchmark tests using ESM-2 have shown that changing the model size from the 650M, used in this study, to 3B or 15B can lead to slight improvements in performance regarding the prediction of protein structures32. For our method, using larger models like 3B or 15B models may also enhance performance. Recently developed techniques, such as QLoRA, a quantization-based LoRA55, make it possible to fine-tune even 15B models in a limited GPU resource. However, we faced issues with incompatibility between CoVFit and QLoRA, leading us to abandon the development of a QLoRA-based model. It is also important to note that scaling up the model can significantly increase training and inference times.
The fourth consideration is data augmentation for fitness data. In viral genome surveillance, there can be a delay of several weeks to months between the date of sample collection and the date of data submission to databases. Furthermore, in viral genome surveillance, the number of viral genomes newly sequenced have gradually decreased in recent years (https://gisaid.org/). Consequently, the most recent genomic data tends to be under-sampled. However, this recent data is considered to contain more critical information for predicting future variants compared to older data. Therefore, increasing the proportion of recent data in the training dataset or employing data augmentation techniques, which artificially expand the dataset, might enhance the model’s ability to generalize to future variants.
Lastly, since we have not conducted an exhaustive investigation of the model’s hyperparameters, the model’s performance could be improved by adjusting them. Adjustable hyperparameters include data normalization methods, network architecture, task weight balance, the optimizer algorithm, learning rate, and maximum epoch numbers.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.